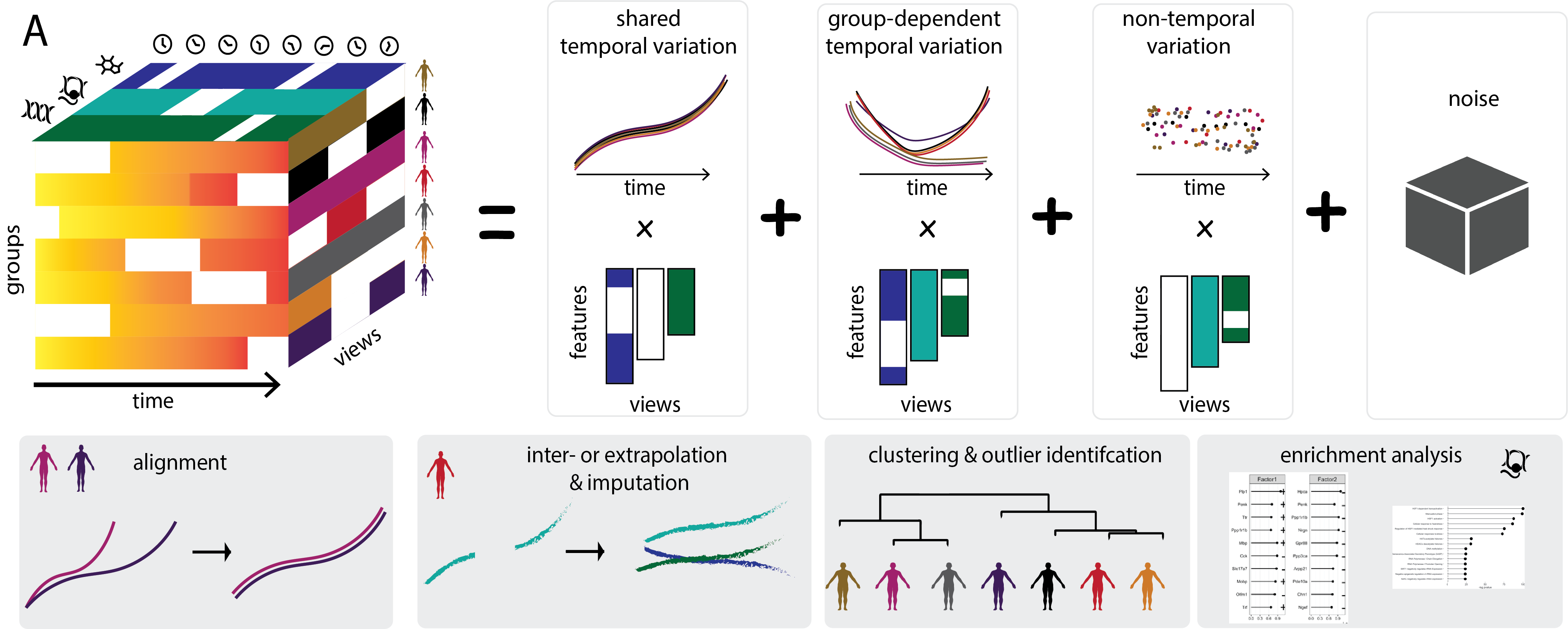
MEFISTO provides an unsupervised approach to integrate multi-modal data with continuous structures among the samples, e.g. given by spatial or temporal relationships. The aim of MEFISTO is to exploit such relationships between samples in the dimensionality reduction and disentangle smooth sources of variation given by factors that change gradually along the covariate and other source of variation that are independent of the covariate. Furthermore, it enables to interpolate/extrapolate to unseen timepoints or locations.
If multiple groups of samples are present, e.g. timecourses from different individuals, MEFISTO furthermore models the heterogeneity of the temporal or spatial patterns across groups and allows to align timepoints across the groups in cases where there are no clear or imperfect correspondences between groups.
For more details you can read our paper:
Installation
MEFISTO was integrated into the MOFA framework and you can follow our installation instructions for MOFA, which provides details on the R and Python package installation as well as a Docker image.
Please make sure you have the latest version of MOFA2 (>=1.1.4) and mofapy2 (>=0.5.8) for MEFISTO to work. If you already using MOFA, you might need to update the R and python package, e.g. using
pip install --upgrade mofapy2
and
devtools::install_github("bioFAM/MOFA2", build_opts = c("--no-resave-data --no-build-vignettes"))
You can also install the R package MOFA2 from Bioconductor. Note that MEFISTO requires the use of the development version. If you have no installation of the python package mofapy2 yet, this will also take care to install these dependencies when training your first MOFA object. Otherwise make sure to have the latest version of mofapy2 (>=0.5.8).
BiocManager::install(version='devel')
BiocManager::install("MOFA2")
Tutorials/Vignettes
- MEFISTO with temporal data: illustration of the method with a temporal covariate
- MEFISTO with spatial data: illustration of the method with a spatial covariate
- Application to an evodevo gene expression atlas: gene expression data from 5 species and 5 organs across development
- Application to a longitudinal microbiome data set: microbiome data from 43 children over the first two years of life
- Application to spatial transcriptomics data: spatial transcriptome data of a mouse brain tissue generated using 10x visium
- Application to single-cell multi-omics: RNA expression, DNA methylation and chromatin accessibility profiled from the same cell using scNMT-seq
Some notes and guidelines on data preprocessing for the use of MEFISTO can be found in this vignette.
Python Tutorials
- Application to an evodevo gene expression atlas: gene expression data from 5 species and 5 organs across development
- Application to a longitudinal microbiome data set: microbiome data from 43 children over the first two years of life
- Application to spatial transcritptomics data: spatial transcriptome data of a mouse brain tissue generated using 10x visium
For Python users, we also recommend the use of muon, a framework that eases the handling of multimodal omics data. We provide tutorials for the use of MEFISTO as part of muon here.
FAQ
(1) When should I use the MEFISTO options in MOFA?
MEFISTO can be used if you have metadata on your samples that give information on how samples relate to one another such as temporal or spatial positions. Using such known similarities can improve the inferred factors, provides the ability to interpolate and enables to separate factors that vary smoothly along these known covariates and those that capture variation independent of them. In particular with many missing samples, MOFA without the use of MEFISTO options can have difficulties to detect such smooth sources of variation. By exploiting known relationships between samples, MEFISTO can better infer such smooth variation.
(2) What is the input to MEFISTO?
Along with the omics data for one or multiple omics and one or multiple groups of samples (as in MOFA without the use of MEFISTO options) you now additionally need to provide covariate values for each sample. This can for instance be a single covariate such as a time point per sample or multiple covariates such as x-,y- coordinate of spatial postions. In R this is done by the function set_covariates which is called on an untrained MOFAobject prior to model training.
(3) How does the smooth factor inference work in MEFISTO?
Like in MOFA without the use of MEFISTO options, factors are inferred to represent the driving sources of variation across data modalities. By specifying sample covariates the model can learn factors that vary smoothly along the covariates. Technically, this is implemented using a Gaussian process prior for the factors with a squared exponential kernel in the covariates. For each factor the model learns a different scale parameter: For factors that vary smoothly with the covariate this will be close to 1, factors that capture variation independent of the covariate will have a scale close to 0.
(4) What if I have distinct groups of samples? Can I specify distinct sample groups in MEFISTO?
Yes you can. This can be very useful, if you have multiple repeated measurement on spatial or temporal data, e.g. time course data from multiple individuals. You can specify the groups by providing the group label for each sample when creating the MOFAobject (see for example the evodevo tutorial). MEFISTO will then infer latent factors for each group and (if model_groups is set to True) infer a group-group correlation matrix that indicates for each factor how the groups relate to one another. See also our evodevo tutorial for an example. Note that setting model_groups to True can be slow for large number of groups - in this case, we recommend setting it to False for initial analysis.
(5) What if my covariates are not aligned across groups?
If you have muliple groups where the covariate is not aligned, e.g. time course data across development from different species, MEFISTO provides an option to learn an optimal alignment. For this, you can use the warping option. See also our evodevo tutorial for an example of the alignment functionality of MEFISTO. Note that for this you need to choose a reference group to which to align the samples. If you want to align time points a higher level than the groups, e.g. aligning individuals from one species or disease type to another but not aligning within the species or disease type, you can specify additional assignments of groups to warping_groups.