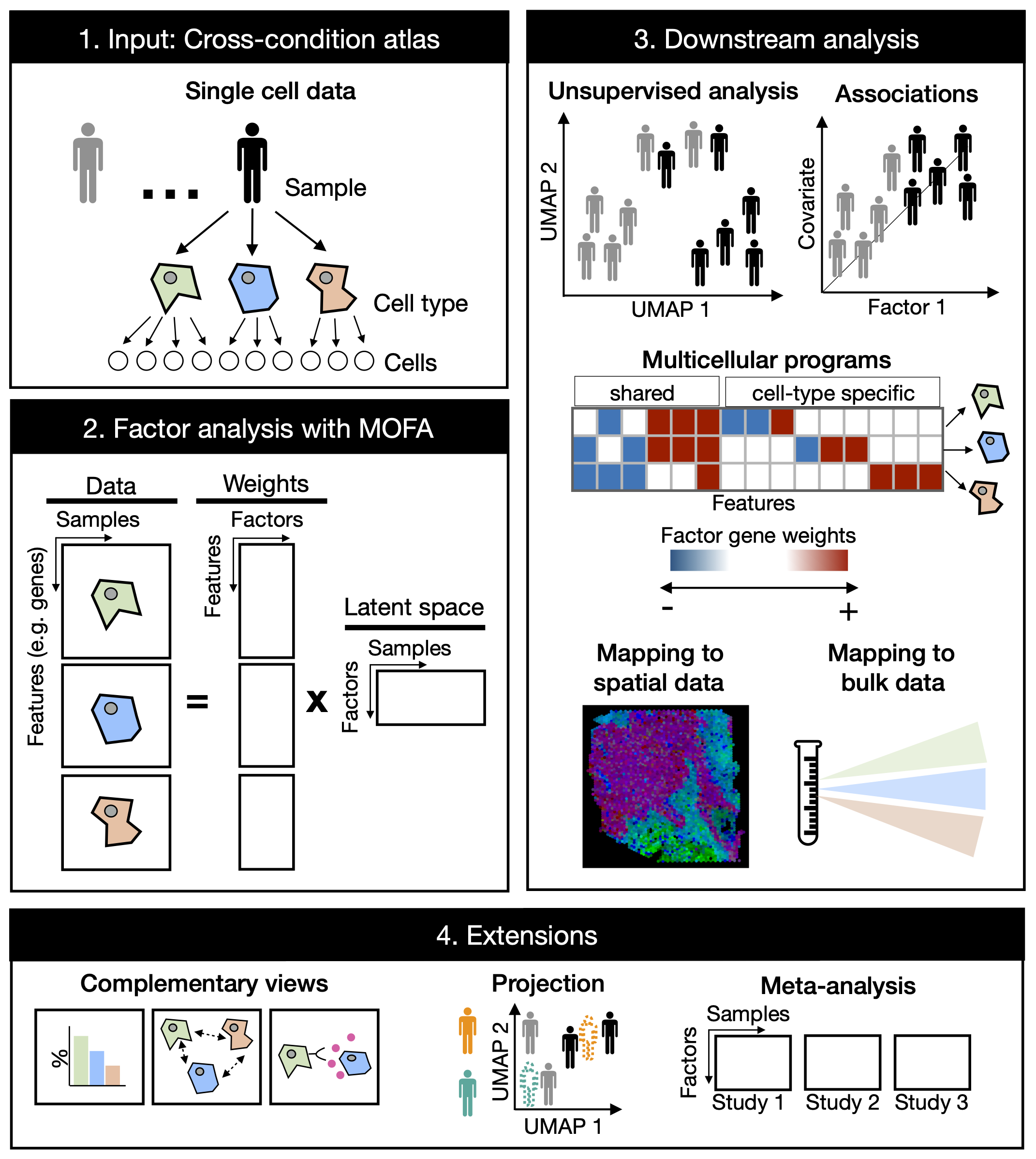
Cross-condition single-cell omics data profile the variability of cells across cell-types, patients, and conditions. Multicellular factor analysis (MOFAcell) repurposes MOFA to estimate cross-condition multicellular programs from single-cell data. These multicellular programs represent coordinated molecular changes occurring in multiple cells and can be used for the unsupervised analysis of samples in single-cell data of multiple samples and conditions. The flexibility in view creation allows the inclusion of structural (eg. spatial dependencies) or communication tissue-level views in the inference of multicellular programs. Leveraging on MOFA’s structured regularization MOFAcell is also suitable for meta-analysis and the joint modeling of independent studies.
For more details you can read our paper: \n
Use
We have created a complementary R package MOFAcellulaR that contains helper fuctions to prepare your single-cell data for a multicellular factor analysis with MOFA.
A python implementation with muon is available through liana-py
Tutorials/Vignettes
- Running a multicellular factor analysis in a cross-condition single-cell atlas: illustration of the method with a toy example
Python Tutorials
- Running a multicellular factor analysis in a cross-condition single-cell atlas: illustration of the method with real data
- Multicellular factor analysis for intercellular context factorization: inference of multicellular programs from cell-cell communication inference scores