Overview
MOFA is a factor analysis model that provides a general framework for the integration of multi-omic data sets in an unsupervised fashion. Intuitively, MOFA can be viewed as a versatile and statistically rigorous generalization of principal component analysis to multi-omics data. Given several data matrices with measurements of multiple -omics data types on the same or on overlapping sets of samples, MOFA infers an interpretable low-dimensional representation in terms of a few latent factors. These learnt factors represent the driving sources of variation across data modalities, thus facilitating the identification of cellular states or disease subgroups.
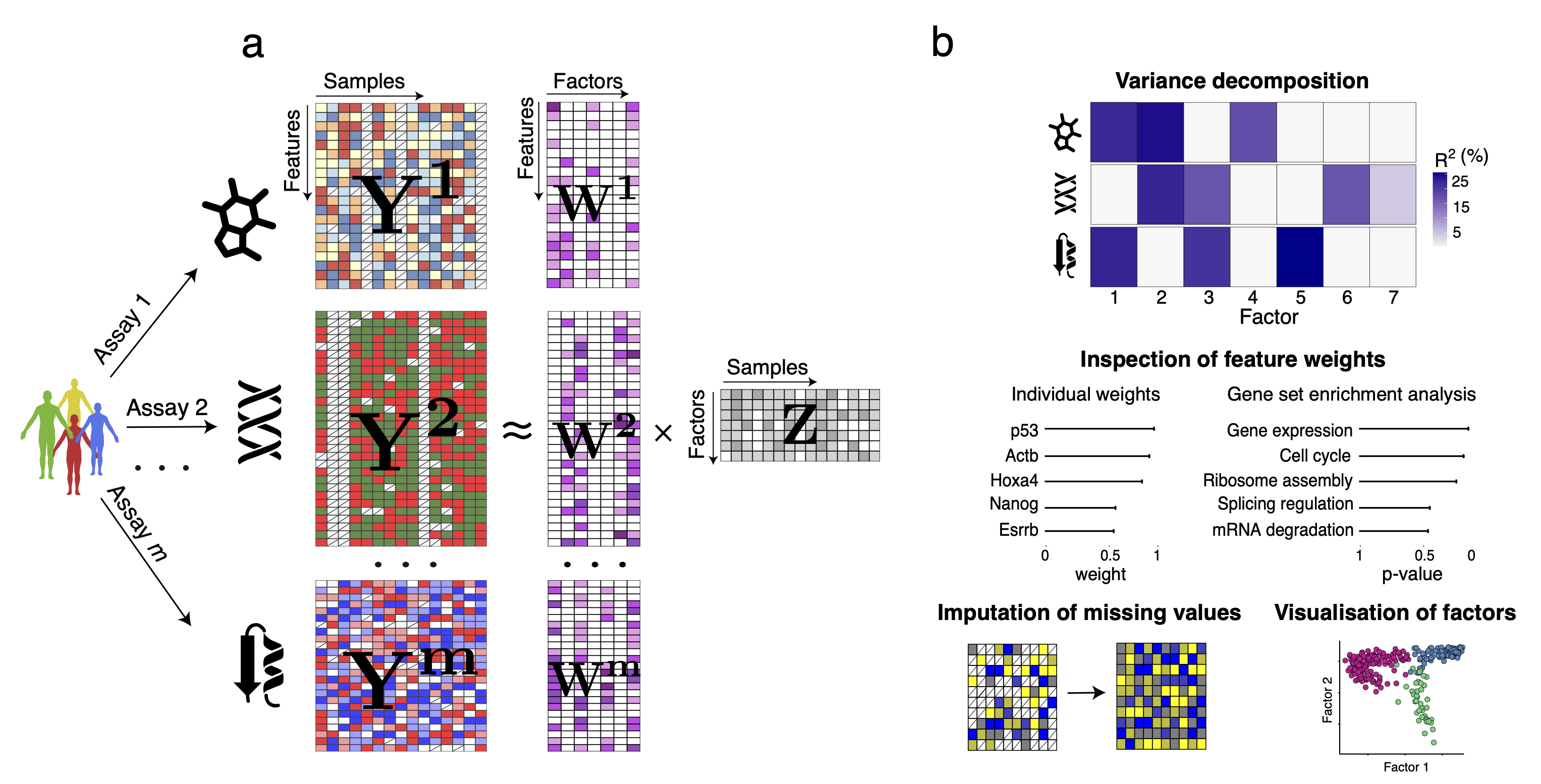
For more details you can read our papers:
- general framework: published in Molecular Systems Biology
- multi-group framework and single cell applications: MOFA+, published in Genome Biology
- temporal or spatial data: MEFISTO, published in Nature Methods
- multicellular factor analysis MOFAcell, published in eLife
Implementation
MOFA is implemented in Python (mofapy2) and R (MOFA2). See installation instructions. Previous implementations of MOFA (mofapy and MOFA) are deprecated and no longer maintained. See News for an overview of changes in the most recent version of the implemenation and a comparison to older implementations.
Citation
If you have used MOFA, please consider citing any of the two articles:
@article{Argelaguet2018,
author = {Argelaguet, R. and Velten, B. and Arnol, D. and Dietrich, S. and Zenz, T. and Marioni, J. C. and Buettner, F. and Huber, W. and Stegle, O.},
title = {Multi-Omics Factor Analysis-a framework for unsupervised integration of multi-omics data sets},
journal = {Mol Syst Biol},
year = {2018},
volume = {14},
number = {6},
pages = {e8124}
}
@article{Argelaguet2020,
author = {Argelaguet, R. and Arnol, D. and Bredikhin, D. and Deloro, Y. and Velten, B. and Marioni, J.C. and Stegle, O.},
title = {MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data},
journal = {Genome Biology},
year = {2020},
volume = {21},
number = {1},
pages = {111}
}
If you have used MEFISTO, please consider citing the following article:
@article{Velten2020,
author = {Velten, B. and Braunger, J.M. and Arnol, D. and Argelaguet, R. and Stegle, O.},
title = {Identifying temporal and spatial patterns of variation from multi-modal data using MEFISTO},
journal = {bioRxiv},
year = {2020},
url = {https://www.biorxiv.org/content/10.1101/2020.11.03.366674v1}
}